This python script is capable of generating a set of GAMESS input files for potential energy surface scanning. It needs a reference GAMESS input and XML file specifying degrees of freedom to scan. PES scan can be done using linear translations, and rotations of atomic groups. Translation vectors can be specified using pair of atoms (bonds) or raw vectors. Axis of rotation can be specified using raw vectors, triplets of atoms (plane angle scan) or pair of atoms (rotation around bond scan).
To use the script save two files «vector.py» and «pes-scan.py» in the same directory. At least two input files are consumed by the script. The first one is a reference GAMESS input which will be replicated by substituting respective coordinates to it. Here is one, defining propane molecule:
$CONTRL COORD=CART UNITS=ANGS $END $DATA test.pdb C1 C 6.0 -0.0000000000 -0.0000000000 0.0000000000 C 6.0 1.1710000000 0.9750000000 -0.2200000000 C 6.0 2.2660000000 0.2830000000 -1.0520000000 H 1.0 -0.7850000000 0.2440000000 -0.6530000000 H 1.0 0.3220000000 -0.9810000000 -0.1900000000 H 1.0 1.5670000000 1.2620000000 0.7100000000 H 1.0 2.6940000000 0.9760000000 -1.7140000000 H 1.0 -0.3350000000 0.0730000000 0.9930000000 H 1.0 0.8280000000 1.8250000000 -0.7310000000 H 1.0 1.8430000000 -0.5080000000 -1.5980000000 H 1.0 3.0080000000 -0.0920000000 -0.4110000000 $END
The second one is an XML, specifying degrees of freedom to scan.
Here is sample XML, specifying 2D PES scan. One coord is torsion angle of terminal methyl, the second is C-C bond length of terminal methyl:
<scan> <coord type="torsion" start="-180.0" stop="180.0" nsteps="12"> <atoms>2 1</atoms> <move>1 4 5 8</move> </coord> <coord type="shift" start="0" stop="1.0" nsteps="2"> <atoms>2 1</atoms> <move>1 4 5 8</move> </coord> </scan>
Coordinate type can be one of the following: shift, angle or torsion. Respective vector is defined as pair of atoms IDs <atoms>I J</atoms> in case of either bond or torsion, or as three atoms IDs <atoms>I J K</atoms> in case of valence angle. Alternatively translational/rotational vector can be defined as a raw vector using container <vector>X Y Z</vector>. In case of rotation specify also origin of rotation using container <origin>X Y Z<origin>. All shifts are relative reference geometry given in input GAMESS file. Container <move<ID1 ID2 ID3 …<move> specifies atoms to move.
Given these two files run the following command:
$ ./pes-scan.py -p -i test.inp -c test.xml
which will produce a set of «conf_ID.inp» files, which represent single points on a grid to scan. You can convert resulting files to pdb format with OpenBabel and load them with your favorite molecular visualization program to check the resulting geometries:
$ for f in conf*.inp; do babel -igamin $f -opdb `echo $f | sed 's/inp$/pdb/'`; done
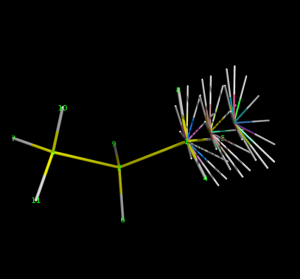
Here is superimposed set of structures, generated using the sample files:
Supporting module «vector.py», needed by main script. Save it and put in the same directory where main script resides.
import math
class vector3:
def __init__(self, x=0, y=0, z=0):
self.x=x
self.y=y
self.z=z
def __add__(self, v):
a = vector3(self.x+v.x, self.y+v.y, self.z+v.z)
return a
def __iadd__(self, v):
self.x+=v.x
self.y+=v.y
self.z+=v.z
return self
def __sub__(self, v):
a = vector3(self.x-v.x, self.y-v.y, self.z-v.z)
return a
def __rmul__(self, c):
a = vector3(self.x*c, self.y*c, self.z*c)
return a
def __imul__(self, c):
self.x*=c
self.y*=c
self.z*=c
return self
def __mul__(self, v):
a = self.x*v.x+self.y*v.y+self.z*v.z
return a
def __pow__(self, v):
a=vector3(self.y*v.z-self.z*v.y, -self.x*v.z+self.z*v.x, self.x*v.y-self.y*v.x)
return a
def norm(self):
l = math.sqrt(self.x**2 + self.y**2 + self.z**2)
return l
def normalize(self):
l = self.norm()
self.x /= l
self.y /= l
self.z /= l
# return self
class matrix3x3:
def __init__(self):
self.m11=1
self.m12=0
self.m13=0
self.m21=0
self.m22=1
self.m23=0
self.m31=0
self.m32=0
self.m33=1
def __mul__(self, v):
a = vector3(self.m11*v.x + self.m12*v.y + self.m13*v.z, self.m21*v.x + self.m22*v.y + self.m23*v.z, self.m31*v.x + self.m32*v.y + self.m33*v.z);
return a
def rot(self, v, t_deg):
t=math.pi*t_deg/180.0
self.m11 = v.x*v.x*(1-math.cos(t))+math.cos(t)
self.m12 = v.x*v.y*(1-math.cos(t))-v.z*math.sin(t)
self.m13 = v.x*v.z*(1-math.cos(t))+v.y*math.sin(t)
self.m21 = v.x*v.y*(1-math.cos(t))+v.z*math.sin(t)
self.m22 = v.y*v.y*(1-math.cos(t))+math.cos(t)
self.m23 = v.y*v.z*(1-math.cos(t))-v.x*math.sin(t)
self.m31 = v.x*v.z*(1-math.cos(t))-v.y*math.sin(t)
self.m32 = v.y*v.z*(1-math.cos(t))+v.x*math.sin(t)
self.m33 = v.z*v.z*(1-math.cos(t))+math.cos(t)
Main script «pes-scan.py»
#!/usr/bin/python
####################################
# Potential energy surface scanner #
# by piton at erg.biophys.msu.ru #
####################################
import os, sys, string, re, copy, threading, xml.dom.minidom
from stat import *
from optparse import OptionParser
from vector import *
VALID_COORD_TYPES=["shift", "angle", "torsion"]
global sem, points_done, pd_lock, DEBUG, parse_only
def getText(nodelist):
rc = ""
for node in nodelist:
if node.nodeType == node.TEXT_NODE:
rc = rc + node.data
return rc
class cScanDim:
def __init__(self, type, start, stop, nsteps, move, vector = None, origin = None, atoms = None):
if not type in VALID_COORD_TYPES: raise NameError, "Unknown coord type '"+type+"'"
self.type=type
if not ( (vector and origin) or atoms): raise NameError, "Either atoms or vector and origin should be defined"
self.atoms=[]
if vector and origin:
self.origin=copy.deepcopy(origin)
self.vector=copy.deepcopy(vector)
else:
for iat in atoms.split():
self.atoms.append(int(iat)-1)
if ( len(self.atoms) != 2 and ( type == "shift" or type == "torsion" ) ) or (len(self.atoms) != 3 and type == "angle"): raise NameError, "Inconsistent coord type number of atoms"
self.start=start
self.stop=stop
self.step=(self.stop-self.start)/nsteps
self.vals=[]
i=0
while i < nsteps:
self.vals.append(self.start + i*self.step)
i+=1
self.vals.append(self.stop)
self.move=[]
for at in move.split():
self.move.append(int(at)-1)
def shift(self, mol, value):
if len(self.atoms):
if self.type == "angle":
self.origin = mol.atoms[self.atoms[1]].vector
self.vector = ( mol.atoms[self.atoms[0]].vector - self.origin ) ** ( mol.atoms[self.atoms[2]].vector - self.origin )
else:
self.origin = mol.atoms[self.atoms[0]].vector
self.vector = mol.atoms[self.atoms[1]].vector - self.origin
self.vector.normalize()
if self.type == "shift":
for iat in self.move:
mol.atoms[iat].vector += value * self.vector
else:
M = matrix3x3()
M.rot(self.vector, value)
for iat in self.move:
mol.atoms[iat].vector = self.origin + M * ( mol.atoms[iat].vector - self.origin)
return mol
class cAtom:
def __init__(self, name, znuc, x, y, z):
self.name=name
self.znuc=znuc
self.vector=vector3(x,y,z)
self.basis = ""
class cMolecule:
def __init__(self):
self.atoms=[]
def AddAtom(self, name, znuc, x, y, z):
self.atoms.append( cAtom( name, znuc, x, y, z ) )
def thread_func(inpname, outname, point):
global sem, points_done, pd_lock, parse_only
if not parse_only:
os.system('gamess -i '+inpname+' -o '+outname)
if S_ISREG(os.stat(outname)[ST_MODE]):
re_E = re.compile('^\s*FINAL\s+ENERGY\s+IS\s+(-?\d+\.\d+)\s+AFTER\s+\d+\s+ITERATIONS\s*$', re.IGNORECASE)
out = open(outname, 'r')
for line in out:
res = re_E.search(line)
if res:
point.append(float(res.group(1)))
out.close()
pd_lock.acquire()
points_done+=1
pd_lock.release()
sem.release()
######################
# Parse command line #
######################
parser = OptionParser()
parser.add_option('-i', dest='inp_file', type='string', help='GAMESS input file', metavar='FILE')
parser.add_option('-c',dest='coord_file', help='file with coodinates to scan', metavar='FILE')
parser.add_option('-o',dest='pes_file', help='output file', metavar='FILE')
parser.add_option('-n',dest='np', help='number of processes to spawn', metavar='NP')
parser.add_option('-p', dest='pretend', help='pretend, i.e. just generate input files', action='store_true',default=False)
parser.add_option('-d','--debug', dest='debug', help='Enable extra output for debugging', action='store_true',default=False)
parser.add_option('-l', dest='parse_only', help='only parse log files', action='store_true',default=False)
parser.add_option('-s', dest='sync', help='vary coordinates synchronously', action='store_true',default=False)
(opts, args) = parser.parse_args()
parse_only = opts.parse_only
if not opts.inp_file and not parse_only:
print 'Please specify GAMESS input file'
sys.exit(1)
else: INP_FILE=opts.inp_file
if not opts.coord_file:
print 'Please specify file with coordinates definitions'
sys.exit(1)
else: COORD_FILE=opts.coord_file
if not opts.pes_file and not opts.pretend:
print 'Please specify output file'
sys.exit(1)
else: PES_FILE=opts.pes_file
if not opts.np:
# print 'Number of processes to run is not defined. Will do one point at a time.'
NP=1
else: NP=int(opts.np)
if NP<1: NP=1
DEBUG=opts.debug
#sys.exit(0)
#################
# Read molecule #
#################
print "\nParsing input file..."
sys.stdout.flush()
f=open(INP_FILE, 'r')
indata=0;
gamess_stuff=""
re_DATA = re.compile('^\s*\$DATA\s*$', re.IGNORECASE)
re_END = re.compile('^\s*\$END\s*$', re.IGNORECASE)
re_CART = re.compile('^\s*([^\s]+)\s+(\d+)\.?0\s+(-?\d+\.\d+)\s+(-?\d+\.\d+)\s+(-?\d+\.\d+)\s*$')
mol = cMolecule()
iat=0
for line in f:
if re_DATA.match(line): indata=1
if indata:
res=re_CART.search(line)
if res:
mol.AddAtom(res.group(1), int(res.group(2)), float(res.group(3)), float(res.group(4)), float(res.group(5)))
iat += 1
elif iat and not re_END.match(line):
mol.atoms[iat-1].basis += line
else:
gamess_stuff = gamess_stuff + line
if re_END.match(line): indata=0
f.close()
if DEBUG:
print "\nAtoms:"
i=1
for atom in mol.atoms:
print str(i) + ': ' + atom.name + ' ' + str(atom.znuc) + ' ' + str(atom.vector.x) + ' ' + str(atom.vector.y) + ' ' + str(atom.vector.z)
i+=1
print '\n'
#print gamess_stuff
#for at in mol.atoms:
# print at.name, at.znuc, at.vector.x, at.vector.y, at.vector.z
###########################
# Generate list of coords #
###########################
print "\nParsing coodinates 2 scan..."
sys.stdout.flush()
C=[]
dom_top = xml.dom.minidom.parse(COORD_FILE)
dom_coords = dom_top.getElementsByTagName("coord");
for dom_coord in dom_coords:
coord_type = dom_coord.getAttribute("type")
start_value = float(dom_coord.getAttribute("start"))
stop_value = float(dom_coord.getAttribute("stop"))
nsteps_value = int(dom_coord.getAttribute("nsteps"))
dom_move = dom_coord.getElementsByTagName("move")
if ( len(dom_move) != 1 ): raise NameError, "Multiple or none 'move' containers"
move_list = getText(dom_move[0].childNodes)
dom_atoms = dom_coord.getElementsByTagName("atoms")
if len(dom_atoms):
if len(dom_atoms) != 1: raise NameError, "Multiple 'atoms' containers"
C.append( cScanDim(type=coord_type, start=start_value , stop=stop_value, nsteps=nsteps_value, move=move_list, atoms=getText(dom_atoms[0].childNodes)) )
else:
if coord_type == "angle": raise NameError, "Coord type 'angle' needs to be defined using 'atoms' directive"
dom_vector = dom_coord.getElementsByTagName("vector")
dom_origin = dom_coord.getElementsByTagName("origin")
if ( len(dom_vector) != 1 or len(dom_origin) != 1 ): raise NameError, "Multiple or none 'vector' or 'origin' containers"
text_vector = getText(dom_vector[0].childNodes)
text_origin = getText(dom_origin[0].childNodes)
if ( len(text_vector.split()) != 3 or len(text_origin.split()) != 3 ): raise NameError, "Bad 'vector' or 'origin' definition"
vector = vector3(float(text_vector.split()[0]), float(text_vector.split()[1]), float(text_vector.split()[2]))
origin = vector3(float(text_origin.split()[0]), float(text_origin.split()[1]), float(text_origin.split()[2]))
vector.normalize()
C.append( cScanDim(type=coord_type, start=start_value , stop=stop_value , nsteps=nsteps_value, move=move_list, vector=vector, origin=origin))
# if DEBUG:
# print
# print coord_type, start_value, stop_value, nsteps_value
# print "move_list: ", move_list
# print "vector: ", vector.x, " ",vector.y, " ", vector.z
# print "origin: ", origin.x, " ", origin.y, " ", origin.z
if not len(C): raise NameError, "No coordinates for scan were defined"
#################
# Generate mesh #
#################
print '\n\nGenerating mesh...',
sys.stdout.flush()
if opts.sync:
numpoints = len(C[0].vals)
else:
numpoints=1
for coord in C:
numpoints *= len(coord.vals)
print "Number of points: ", numpoints
mesh=[]
for i in range(0,numpoints):
point=[]
j=0
l=i
while j < len(C):
if opts.sync:
if len(C[j].vals) != numpoints: raise NameError, "Inconsistent number of steps"
point.append(C[j].vals[i])
else:
k = l - l / len(C[j].vals) * len(C[j].vals)
l = l / len(C[j].vals)
point.append(C[j].vals[k])
j+=1
mesh.append(point)
i+=1
print " Done"
########################
# Generate input files #
########################
if not parse_only:
print "\n\nGenerating input files...",
sys.stdout.flush()
iconf=0
for point in mesh:
conf=copy.deepcopy(mol)
i=0
while i < len(point):
conf = C[i].shift(conf, point[i])
i+=1
# write conformation
inpname = 'conf_'+str(iconf).zfill(5)+'.inp'
f=open(inpname, 'w')
f.write('\n $DATA\nconformation #'+str(iconf)+'\nC1\n')
for atom in conf.atoms:
f.write(atom.name+' '+str(atom.znuc)+'.0 '+str(atom.vector.x)+' '+str(atom.vector.y)+' '+str(atom.vector.z)+'\n'+atom.basis)
f.write(' $END\n')
f.write(gamess_stuff)
f.close()
iconf += 1
print " Done"
####################
# Calculate points #
####################
if not opts.pretend:
print "Starting calculations"
sem = threading.Semaphore(NP)
pd_lock = threading.Lock()
points_done=0
thrd_list=[]
for i in range(0, numpoints+NP):
sem.acquire()
print "\rProgress: "+str(points_done)+' / '+str(len(mesh)),
sys.stdout.flush()
if i < numpoints:
inpname = 'conf_'+str(i).zfill(5)+'.inp'
outname = 'conf_'+str(i).zfill(5)+'.log'
thrd_list.append(threading.Thread(target=thread_func, args=(inpname, outname, mesh[i])))
thrd_list[len(thrd_list)-1].start()
for thrd in thrd_list:
thrd.join()
#####################
# Print PES to file #
#####################
log = open(PES_FILE, 'w')
for point in mesh:
for x in point:
log.write(str(x)+' ')
log.write('\n')
log.close()
print "Done"